From predictive to interpretable models
At Volv we provide the insights on our approaches and methodology
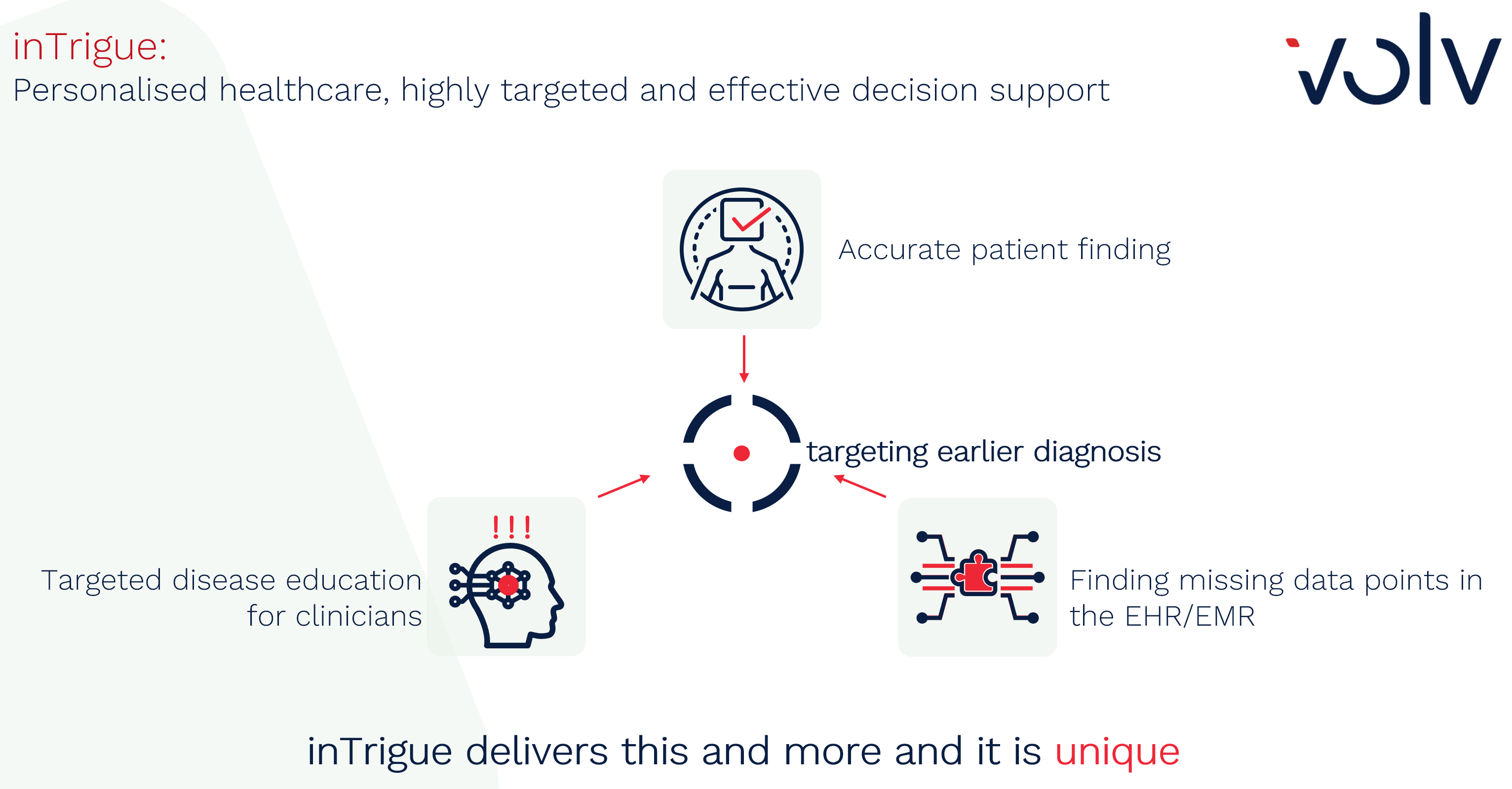
Quantifying Predictive Power of Features in Electronic Health Record Models – the Volv inTrigue way
When we work on complex prediction models with our inTrigue methodology, we are often asked to help clinicians and others to interpret these models by listing the patient features (attributes) which are used by the model to form its predictions. And indeed, generating a list of predictors ranked by their ‘importance’ in a model can translate to improved interpretability and clinical impact. However, there is some work that needs to be carefully considered in order to produce tooling that is derived from complex models that can provide real benefit in a clinical situation. So Rich Colbaugh and I decided to discuss this for you, our audience.
as a challenge for customers and is discussed at many conferences. As it is important to deliver real world results, we discuss here what Volv does to create interpretable models of true utility. The journey from complex modelling to interpretable models is however not necessarily simple when dealing with real-world solutions involving highly dimensional messy data.
Standard feature importance ranking
It is straightforward and often useful to identify features with predictive power, and to quantify their importance, in standard predictive modelling problems. For example, conventional models (e.g., random forests, L1-regularized logistic regression) applied to typical datasets (reasonable dimensionality and noise-levels, ‘vanilla’ correlations) admit this sort of analysis. But there are different considerations to take into account as modelling becomes more complex.
Considerations in feature ranking for EHRs
If we consider predictive modelling for patients based on their GP EHRs, this is now no longer a standard problem, and one should proceed with caution when attempting to quantify the importance of predictive features in this setting. This is because there are specific considerations and characteristics of EHR modelling that Volv have worked with that we would recommend taking into account.
For example, many EHR features are related to each other in a hierarchical manner (e.g., diagnostic codes and medication codes), and this actually represents a more subtle and potentially problematic issue than simple correlation between features. On the other hand, many feature values in patient records are missing (e.g., lab test results in GP EHRs), and therefore, model learning and deployment must be robust to missing features in order to be clinically useful. One also might want to take into account the fact that the ‘labels’ assigned to the examples used to train the models are unreliable, so model learning must be robust to this ‘noise’ as well.
This has a significant implication, as it means that standard methods for quantifying feature importance, such as forward-selection, backward-selection, LASSO-regularization, or classical statistical tests, can produce highly misleading assessments.
Adverse outcomes with feature ranking
One of the things that can go wrong in this scenario is that when applying one of the above techniques to hierarchically organized feature sets (e.g., ICD or ATC codes) it will frequently result in two undesirable outcomes. Firstly, one might achieve unacceptably high sensitivity of feature importance values to variation in the training data, and at the same time the method will often assign low importance to genuinely predictive features; there is also susceptibility to other issues which further reduce dependability and ultimately the clinical utility of the assessments.
Also, to consider is the fact that applying standard methods to models that are robust to missing feature values, which is an essential attribute of practically useful models, then conversely can tend to underestimate feature importance. As we need to provide robustness to label noise, which is also essential for real-world deployment, this will often lead to the discovery and integration of ‘meta-features’ for example patient membership in a particular cluster of other patients, LDA topics in clinical notes, particular sequences of codes. Such features can improve performance substantially, but they also tend to reduce the interpretability and validity of standard estimates of feature importance.
Solutions to feature ranking in complex models
What does Volv do for our customers in these real-world situations? Volv could pick from three broad approaches:
- learn a standard, but not very good, prediction model on a subset of features (ignoring codes, say) and then quantify the importance of that model’s features. This is not recommended, of course, because features which are predictive in a not-very-predictive model may not be predictive for the phenomenon under study. Surprisingly, though, this is done a lot!
- learn a good prediction model and ignore the above concerns we have mentioned – this is not recommended either but is also done a fair amount.
- learn a good/robust prediction model and characterize the predictive importance of the model’s features using a semi-quantitative scheme (e.g., heat-map with a small number of ‘bins’).
Volv inTrigue - interpretive modelling
Or alternatively, what we do at Volv for our customers is to learn an interpretable model from the predictions of a good/robust model (which is proprietary to Volv) and then assess the predictive importance of the features of this new, interpretable model. What that does is deliver a model that can be utilised by a clinician for example, as it is developed in their language and terminology, and it has quantifiable predictive performance. And importantly, we as humans can learn new things from these models that are novel.